Protein Subcellular Localization Prediction
|
|
High throughput proteome sequencing projects continue to churn out enormous amounts of raw sequence data. However, most of this raw sequence data is unannotated and hence, not very useful. Among the various approaches to decipher the function of a protein, one is to determine its localization Experimental approaches for proteome annotation including determination of a protein's subcellular localization are very costly and labor intensive. Besides the available experimental methods, in silico methods present alternative approaches to accomplish this task. Here, we present three in silico machine learning approaches for prediction of the subcellular localization of a protein from the primary sequence information.
Machine learning algorithms, k Nearest Neighbor (k-NN) and Probabilistic Neural Network
(PNN) along with a kernel based Support vector machines (SVM) technique were used to
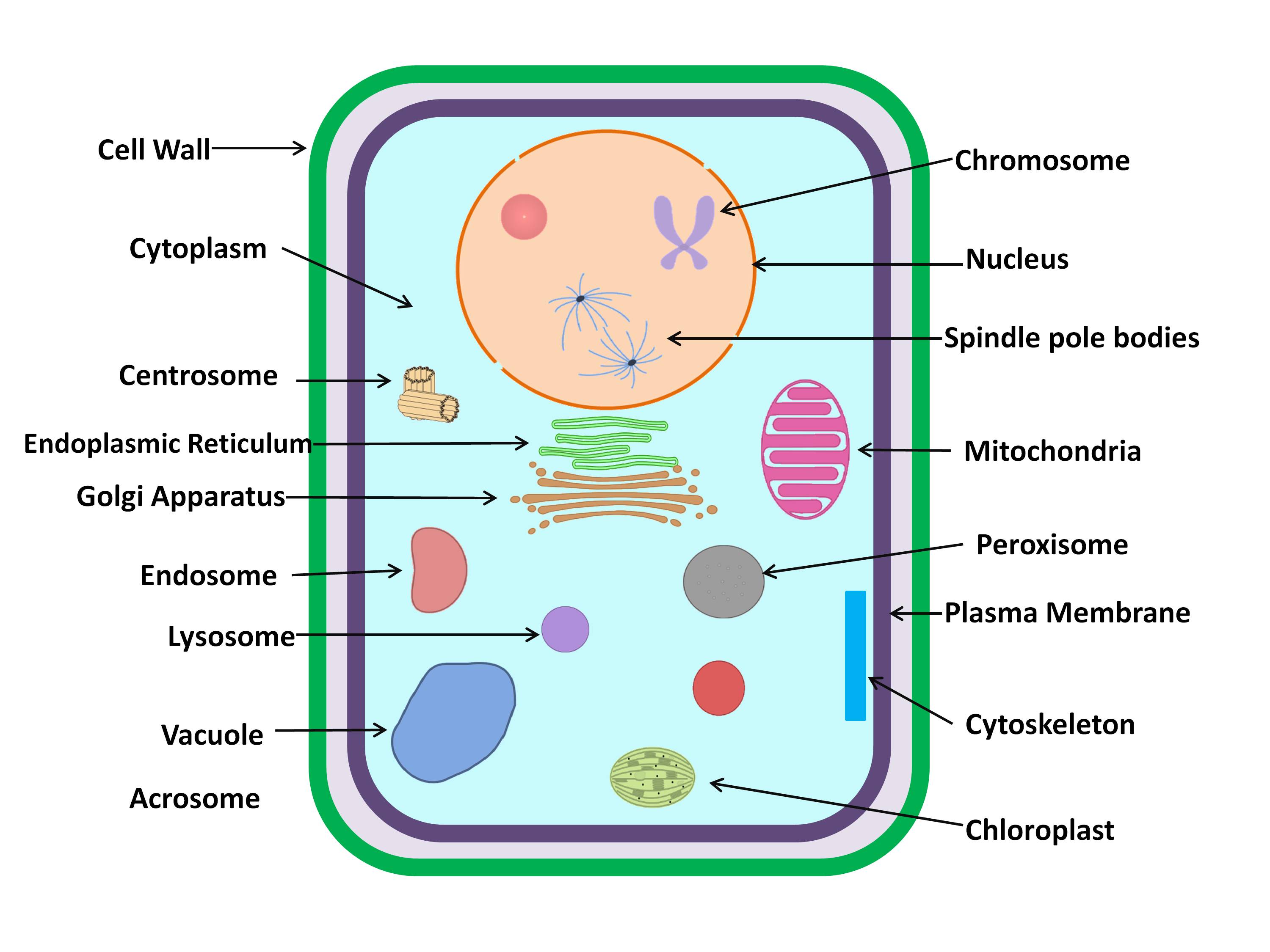
classify an unknown protein into one of the 19 (Acrosome, Cell wall, Centriole,
Chloroplast, Cyanelle, Cytoplasm, Cytoskeleton, Endoplasmic reticulum, Endosome,
Extracellular, Golgi apparatus, Lysosome, Mitochondria, Nucleus, Peroxisome, Plasma
membrane, spindle pole body, Synapse and Vacuole) subcellular localizations. The final
prediction is made on the basis of a consensus of the predictions made by selected algorithm and a probability is assigned to it.