Algorithms
After collecting the data, data mining algorithms are used to extract useful information from the data. Data mining algorithms,Artificial Intelligence (AI) techniques and support vector machine algorithm are used to assign subcellular location to the "Hypothetical" proteins. In this work Probabilistic Neural Network (PNN), k-Nearest Neighbor (kNN) and Support vector machines (SVM) are used to make predictions for the subcellular localization.
k-Nearest Neighbor (k-NN)
In pattern recognition, the k-nearest neighbor algorithm is a method for classifying objects based on closest training examples in the feature space. k-NN is a type of instance-based learning where the function is only approximated locally and all computation is deferred until classification. To implement the k-NN algorithm on any classification problem, a labeled dataset and a metric to measure the proximity of the two vectors in an n-dimensional space is required. For each protein, its dot product with all other proteins in the dataset will be calculated. Based on this dot product, its cosine with any other protein in the n-dimensional space is calculated as given by Equation:
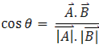
This cos_theta metric will be used to measure the proximity of a protein to another protein in the space. Based on this metric, the nearest neighbor(s) for an unknown object is identified. The class of the nearest neighbor(s) is then assigned to the unknown object. When only one closest neighbor is taken into consideration, the algorithm is known as Nearest Neighbor algorithm. In case, more than one neighbor is considered, the class of the unknown object is assigned by a majority voting scheme. Greater is the value of cos θ between two proteins, closer are the two proteins. For proteins which have a cos θ value of 1, the two proteins exactly super-impose on each other in the space. In this study we have used the value of k=1.
Probabilistic Neural Networks (PNN)PNN are conceptually similar to k-NN models. The k-NN algorithm only considers the nearest object (neighbor) to an unknown object in the domain space and assigns a class to the unknown object, while the PNN algorithm considers all training examples in determining the final class of the unknown object. In the first layer, the distance is computed from the point being evaluated to each of the other points, and a radial basis function (RBF) (also called a kernel function) is applied to the distance to compute the weight (influence) for each point. The radial basis function is so named because the radius distance is the argument to the function. The second layer sums these contributions for each class of inputs to produce as its net output a vector of probabilities. Finally, a compete transfer function on the output of the second layer picks the maximum of these probabilities, and produces a 1 for that class and a 0 for the other classes.
These two algorithms are used to predict subcellular localization. The idea of using two algorithms is to consider the local and global effect of prediction. In kNN only nearest neighbors are considered while in PNN all the learning examples are considered for the prediction.Besides these algorithm a kernel based method called support vector machine is also used for prediction.
Support Vector Machines (SVM)SVMs are universal approximators based on statistical and optimization theory. The SVM is particularly attractive to biological sequence analysis due to its ability to handle noise, large dataset and large input spaces. In the present study, we have used LIBSVM to predict the subcellular localization of proteins. The software allows the users to define a number of parameters and also enables a choice of inbuilt kernel function, like linear, RBF and Polynomial. We have used RBF kernel for model generation. Before generating models from training set, a thorough search for two parameters "C" and gamma was carried out. The values for which maximum accuracy obtained after 10 folds cross validation are then used for model generation. The prediction of subcellular localization is a multi-class classification problem. We developed a series of binary classifiers to handle this multi-classification problem. In this way, fifteen SVM models were constructed for various subcellular localization of protein inside cell.
Protein BlastProtein Blast or "blastp" is based on Basic local alignment search tool.It uses a sequence comparison algorithm - a procedure for solving a mathematical problem in a finite number of steps that frequently involves repetition of an operation, optimized for speed used to search sequence databases for optimal local alignments to a query. The BLAST Algorithm is fast and sensitive. Sequences are filtered to remove low complexity regions (to optimize a meaningful sequence alignment). This search directly compares the input protein queries against a protein database. It is useful in finding other proteins that share sequence similarities to the input query. The matching sequences found by blastp can help determine the nearest protein that may share the subcellular localization with that of the input query. Swissprot database is used as standard database. The alignment is carried out with the help of NCBI web service.