 CRAFT
( Cavity
Recognition
Assisted by
Flow
Transfer Algorithm)
CRAFT
( Cavity
Recognition
Assisted by
Flow
Transfer Algorithm)
 CRAFT
( Cavity
Recognition
Assisted by
Flow
Transfer Algorithm)
CRAFT
( Cavity
Recognition
Assisted by
Flow
Transfer Algorithm)
A novel Delaunay-based “Flow transfer algorithm” to address protein cavities with distinct van der Waals radii.
Visualization penal facilitate user with various handy options to analyze cavities along with useful information.
An automated tool that allows users to customize parameters according to their specific requirements.
Accepts standard.pdb files and offers dedicated modules for selecting chains, ligands, cofactors,and alternate positions, while excluding water molecules automatically.
Numerous physicochemical properties such as cavity volume, solvent accessible surface area, donor, acceptor, aromatic ring, enclosure, exposure, number of cavity opening etc. are estimated for each cavity
A graphical representation depicts the individual residue's contribution to cavity formation
All types of interactive cavities such as voids, channels, pockets, and surface cavities are predicted using the CRAFT.
Disclaimer: Avoid selecting ligands unless essential.
Input .pdb file must follow standard PDB file format. For more information about standard PDB format https://www.cgl.ucsf.edu/chimera/docs/UsersGuide/tutorials/pdbintro.html
Hydrogen atoms must be present after non hydrogen atoms of respective residue.
Cavity properties such as donor, acceptor, and aromatic ring do not consider ligand atoms owing to lacks ligand connectivity information in some pdb files.
SASA of ligand or metal atoms is calculated by the guessing their van der Waals radii or if unable to guess then van der Waals radii set to zero.
Default “Extended Boundary” setting peel off bigger and smaller tetrahedrals form the convex hull, but change in setting only removes bigger tetrahedrals.
Default “Extended Forbidden Face” setting terminate algorithm with encounter with forbidden face, but change in settings add neighbor tetrahedral of forbidden face.
Errors like "hetatm" arise due to discontinuous or missing atom numbers in PDB files. To address these errors, ensure the PDB file adheres to the standard format, maintaining continuous and accurate atom numbering.
Delaunay triangulation: Delaunay triangulation, are used to discrete a domain of points into an unstructured mesh network. The 2D Delaunay triangulation (DT) of a given set S of n points is a triangle simplicial decomposition of convex hull of S such that circumcircle of every simplex is empty i.e. it does not contain any point from S in its interior. The DT are unique for a given set of points and each triangle consists of three vertices, and three edges. The DT generates high quality of mesh for finite set S, by maximizing the minimum angle and minimizing the maximum circumsphere radius of all possible triangulations.
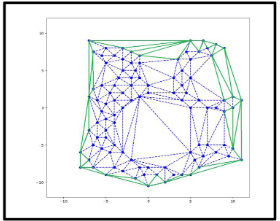
a) Consider 2D points in a plane such that each point has a invisible circle of different radius to represent spherical atoms of a given protein. Euclidean distance between the considered points is used to define bound or unbounded atoms of protein. 2D Delaunay triangulation are represented with triangle simplexes having a regular mesh i.e. without any overlapping between the simplexes.
b) Next step is to find delimiter surface triangle for given point set. A surface triangle can easily be identified from the rest triangles based on its neighboring triangles that would be less than 3. These surface triangles are further peel off until they and their neighbors have circumcircle greater than peeling off limit. Atoms of all these triangles will act as delimiter surface atoms. Therefore, any triangle originating from these atoms will be considered as surface delimiter triangle (represented:in green color)
c) Flow Transfer Algorithm: Algorithm proceeds in three steps in 2D:
i). Initiation step ii). Propagation step iii). Termination step
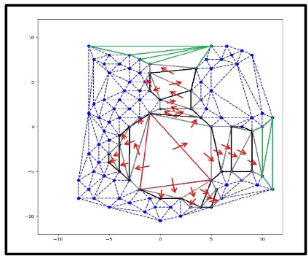
i). Initiation : Algorithm needs a seed triangle per cavity to start search operation. The triangles having greater circumcircle radiuses will be selected from the unstructured Delaunay triangulation mesh, as susceptible triangle. Susceptible triangles are more prone to show their presence in the cavity space and later, act as seed triangle. Here, red triangle is the seed triangle.
ii). Propagation step: The seed triangle flow can only transit through its three edges. The flow transfer from one triangle to other triangle through its mutual edge, only when their mutual edge satisfies the transit/edge condition. The transit/edge condition is defined such that their mutual edge must have enough space to transit to its neighbor, after excluding the radiuses of circles present at its vertexes. The flow transfer is shown with the red arrows.
iii) Termination step: The flow is either terminated by failure of mutual edge transit condition or by the presence of edges of delimiter triangles.
Likewise, a smooth all direction screening will be performed from the seed triangle to lineate the surface atoms of the cavity.
3D Delaunay triangulations are used to implement algorithm for cavity prediction in a given protein. 3D algorithm will use tetrahedral simplexes instead of triangles, seed tetrahedral initiates cavity search and Maximum Circle Radius (MCR) is used instead of edge transit condition. Maximum Circle Radius (MCR) is a modified concept of weighted Delaunay triangulation to consider different van der radii of atoms.
Reference: If the webpage was helpful to your research, please include the following article citation.
1. Open the calculation section on the CRAFT "Home" page.
2. Upload a protein structure file in .pdb format for cavity identification. Customize parameters if required; otherwise, proceed with default settings.
3. Select chains within the pop-up Module and decide whether to include associated ligands.
4. If ligand consideration is chosen and present, select them using another pop-up Module.
5. A last Module activates if chains include alternative amino acid positions; proceed otherwise.
6. Submit chosen chains and ligands to the Flow Transfer algorithm.
7. View identified and characterized protein cavities on the output webpage.

Anuj Gahlawat |

Dr. Prabha Garg
|